Documents for E3Docker
1. Search
In the search box, enter the relevant information of the E3 ligase you are looking for, such as UniProt accession number, gene name, and protein name, for example Q96SW2, CRBN, Cereblon.
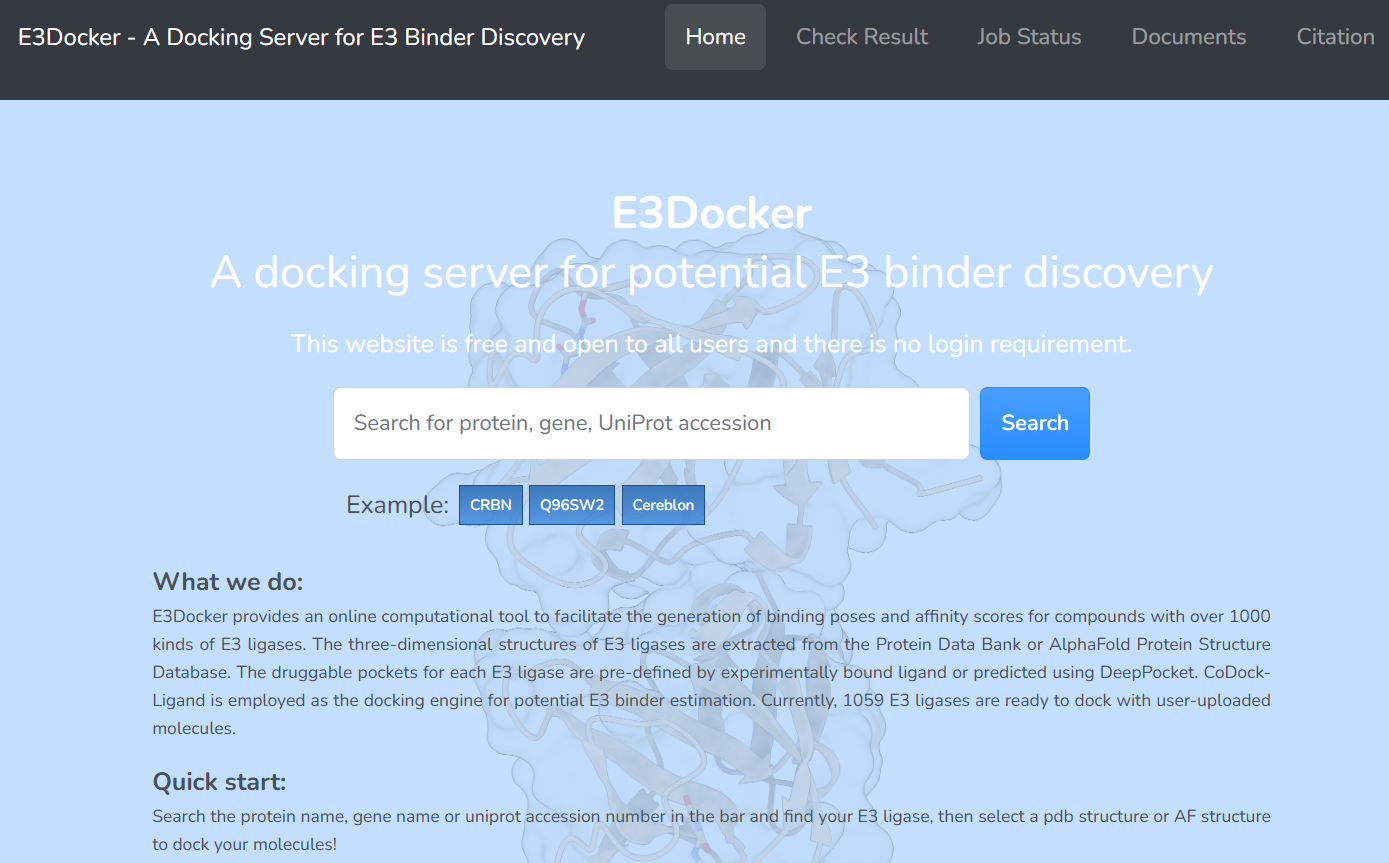
2. Select your protein
On the search results page, we provide a list of proteins that matches your search. Additionally, we offer corresponding information such as Function, Category, Sequence, etc., to assist you in confirming the protein you intend to dock with. After confirming the protein you want to dock, click the “Docking” button to enter the docking page.
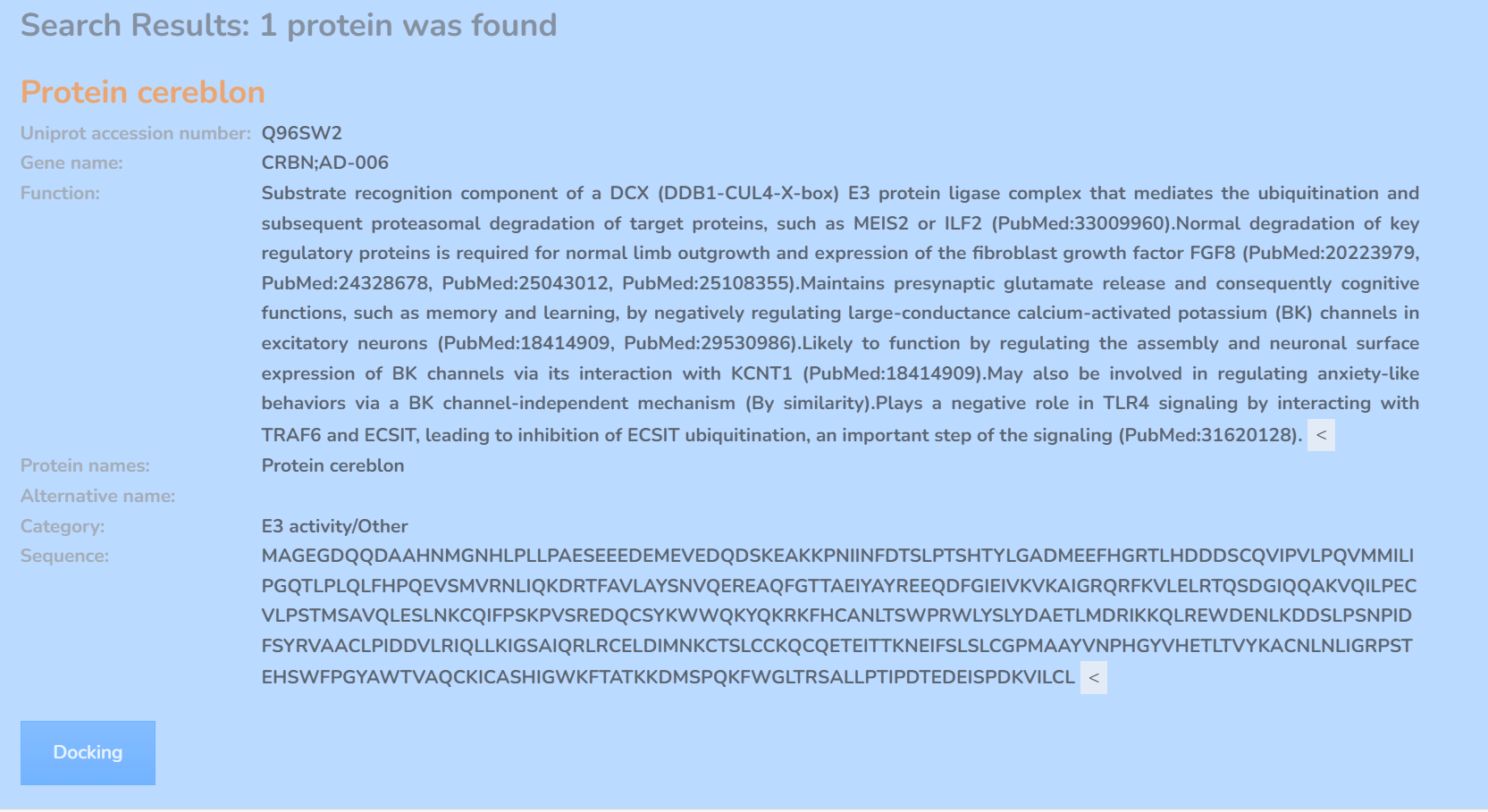
3. Select PDB structures or AlphaFold predicted structure for docking
On the docking page, we provide the PDB structures corresponding to the selected E3 ligase, as well as AlphaFold predicted structure. For the AlphaFold predicted structures, we provide the average confidence score. The druggable pockets for each E3 ligase are defined by experimentally bound ligand from PDB or predicted by using DeepPocket. The docking box is set to the central coordinates of pocket with the size of 24×24×24 Å, making it contains the entire pocket area. You can select the corresponding structure and coordinates of pocket for docking.
We also provide an option for recommended PDB structures on the webpage, specifically designed for non-expert users. The recommended PDBs are selected with highest resolution and in complex with co-crystallized ligands. If no complex form is available, the PDBs with highest resolution and no mutations are chosen. In cases where only an AF structure is available for an E3 ligase, this structure is used as the recommended one. Users can select a structure for docking by clicking its PDB ID.
Binding site determination is a central factor in molecular docking and directly influences the quality and reliability of docking results. The docking boxes are predefined for user selection. For protein-ligand complex PDB structures, the geometric center of the ligand was used to define the binding pocket. For structures without known ligand-binding information, such as apo protein structures from the PDB or predicted protein structures from the AlphaFold Protein Structure Database (AFDB), DeepPocket was employed to identify potential druggable pockets, which were then used as docking sites. For DeepPocket predicted structures, a maximum of three top-scoring pockets are retained. The center of the native ligand or the DeepPocket-defined site was used to set a cubic docking box with dimensions of 24 Å × 24 Å × 24 Å. Furthermore, TM-align is employed to align the highest-resolution ligand-bound experimental structure—designated as the recommended structure on the webpage—with the corresponding apo and AF structures, thereby mapping the experimental ligand-binding sites onto these structures for user selection.
We provide three options for box selection: (1) select single predefined box for docking; (2) select all predefined box for docking; (3) define a use-specified box for docking.
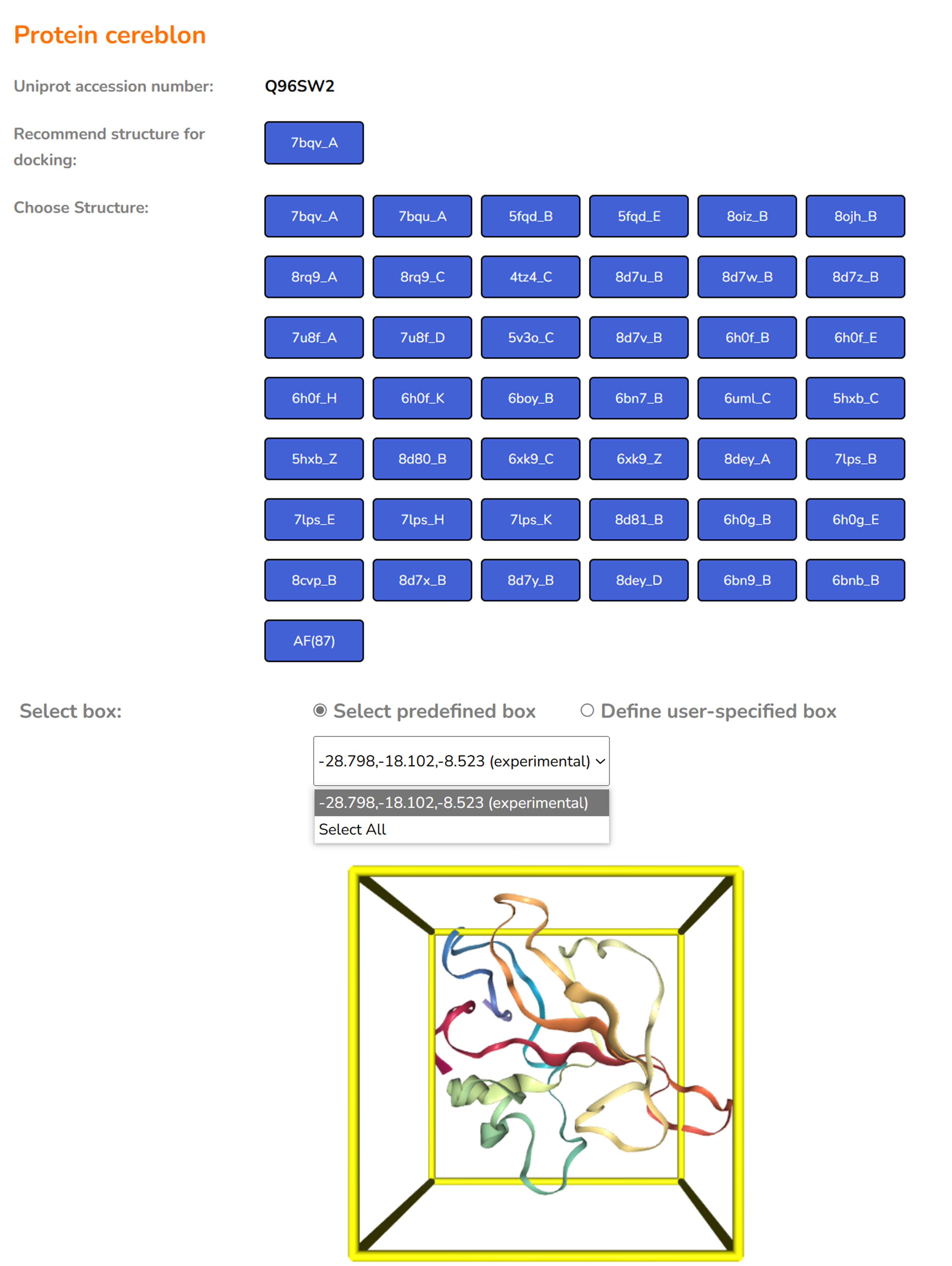
4. Upload ligand file for docking
Please note that you need to upload small molecules in strict mol2, sdf or smi formats. The number of small molecules for batch docking shall not exceed 50. Open Babel ( License: Open Babel Official Page ) is used for format transformation or 3D coordinate generation for the uploaded files. CoDock-Ligand is used as docking engine and it will take a few minutes to finish the docking job. An email will be sent to your email address with a direct link to access your docking results.
5. Retrieve the jobs
After your job is completed, you could enter the job ID to access the result page.
Alternatively, you can access the docking results by the corresponding link in your email. The download url is provided as: http://e3docker.schanglab.org.cn/index/result/index?job_id=******
When the job is completed, the server will send an email to the user’s email address. We use CoDock-Ligand for docking and score ranking. The top 10 models with the lowest scores will be displayed on the results page for your review. The models can be visualized in 3D by NGL (License: NGL). You can view and download your docking results from the result page.
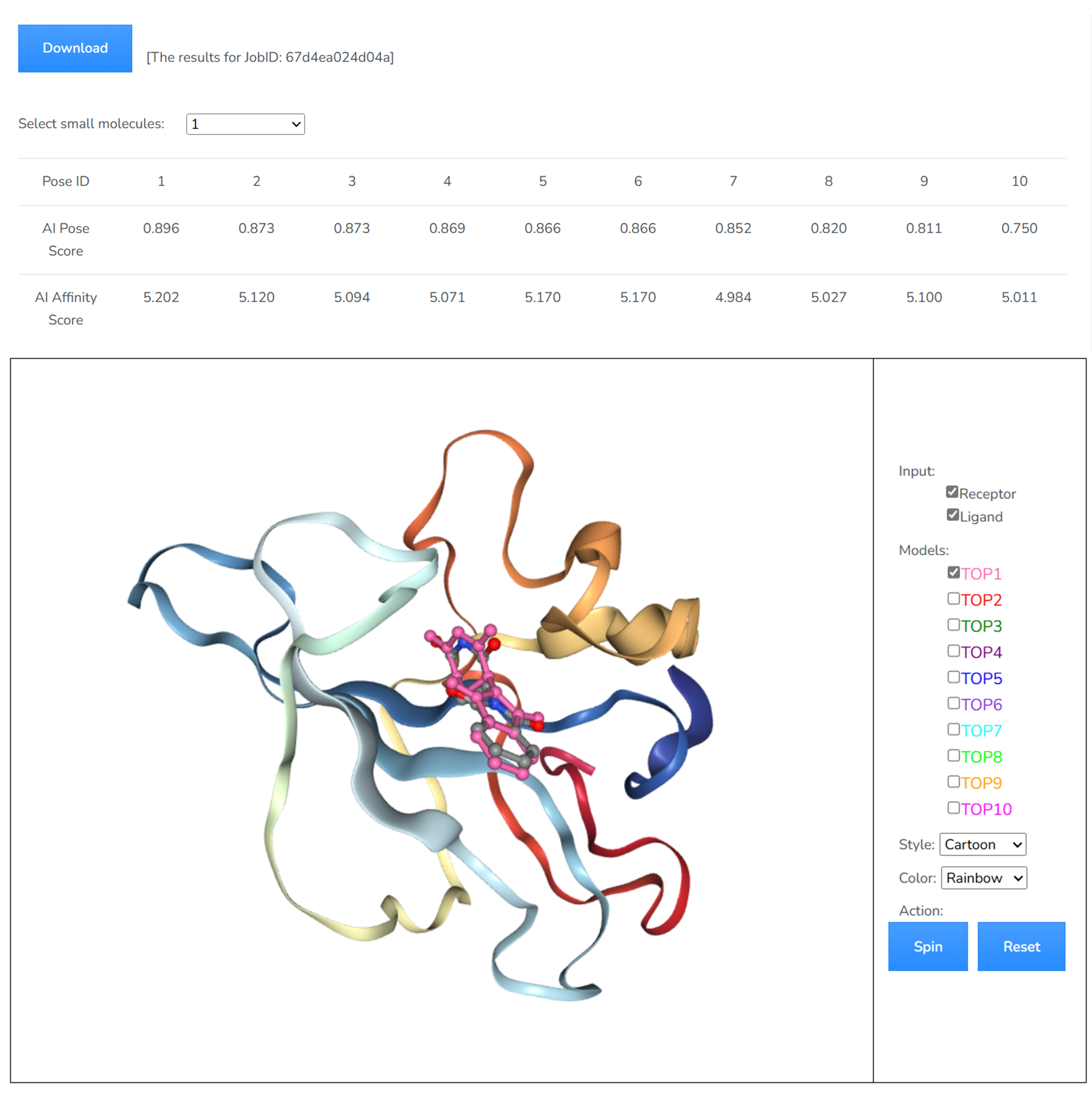
In the downloaded result package, several files are provided. The receptor protein target is renamed as rec.pdb, the uploaded ligand is renamed as raw_1.mol2. The top 10 output ligand poses are named as 0.sdf-9.sdf, respectively. The file of scores.txt contains the score values for each poses.
If you have any suggestions or questions, please Contact Us.